GraphKM: New AI Model to Predict Enzyme Kinetics
A recent study titled “GraphKM: Machine and Deep Learning for KM Prediction of Wildtype and Mutant Enzymes“ has been published in BMC Bioinformatics by researchers Xiao He and Ming Yan from the College of Biotechnology and Pharmaceutical Engineering at Nanjing Tech University, China. This research addresses a significant challenge in enzyme kinetics: accurately predicting the Michaelis constant (KM) for enzymes, which is essential for understanding enzyme behavior but often difficult and resource-intensive to measure experimentally.
Understanding the Michaelis Constant (KM)
The Michaelis constant, KM, is a crucial parameter in enzyme kinetics that represents the substrate concentration at which an enzyme operates at half its maximum velocity. It provides insights into the affinity between an enzyme and its substrate—a lower KM indicates higher affinity. Determining KM values is vital for various biological and biotechnological applications, including drug development, metabolic engineering, and understanding metabolic diseases.
Introducing GraphKM: A Deep Learning Solution
To overcome the challenges of experimentally determining KM, the researchers developed GraphKM, a deep learning model designed to predict KM values for both wildtype (natural) and mutant enzymes. Built on the PaddlePaddle deep learning framework, renowned for its efficiency and scalability, GraphKM integrates advanced computational techniques to enhance prediction accuracy.
Core Components of GraphKM
GraphKM’s architecture combines:
- Graph Neural Networks (GNNs): These networks are adept at handling complex molecular structures by representing molecules as graphs, where atoms are nodes and bonds are edges. This allows the model to capture the intricate details of substrate molecules.
- Transformer-Based Language Model for Proteins: This component processes protein sequences (the enzymes) to understand their biological context and functional properties. Transformers have revolutionized natural language processing and are now being applied to biological sequences to capture long-range dependencies and patterns.
By integrating these two components, GraphKM can analyze both the substrate’s molecular composition and the enzyme’s sequence, providing a comprehensive understanding of their interaction.
Evaluating GraphKM’s Performance
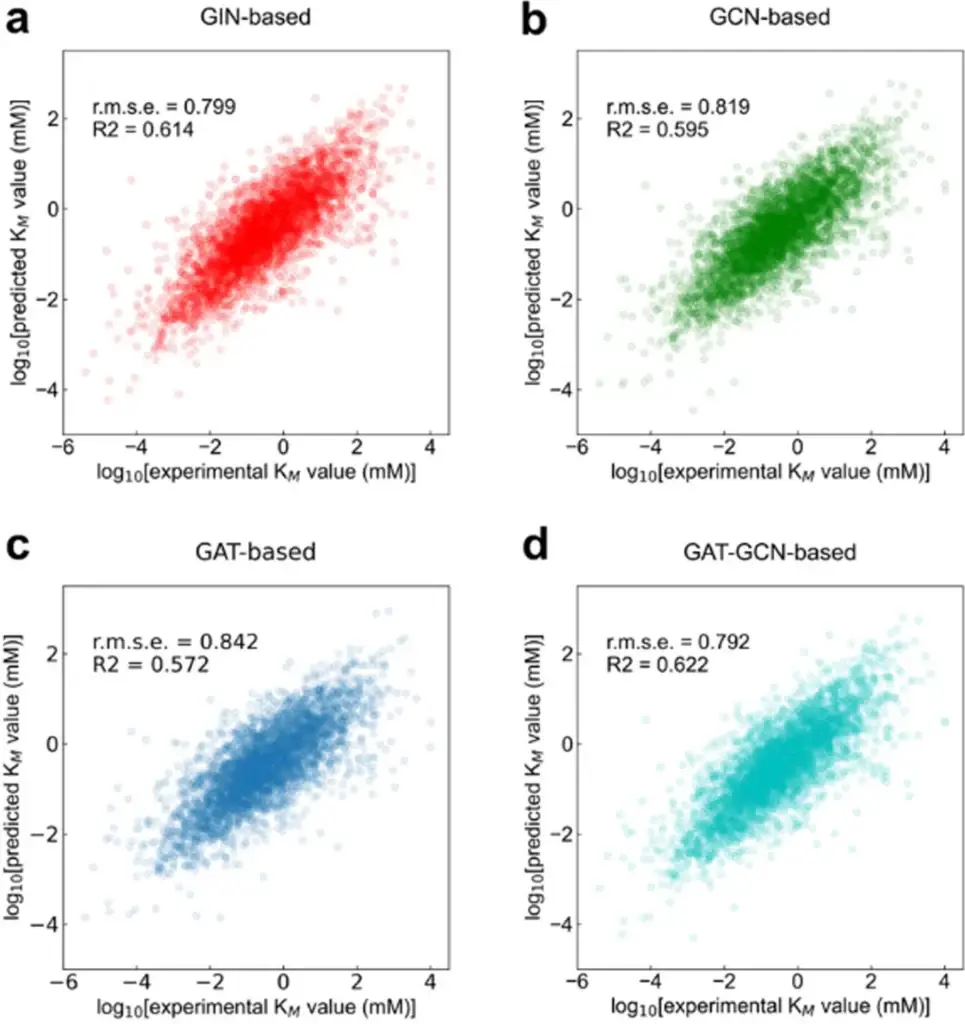
The study evaluated different types of GNNs within the GraphKM framework, including:
- Graph Isomorphism Networks (GIN)
- Graph Attention Networks (GAT)
- Combinations of GNNs (e.g., GAT-GCN)
Among these, the GAT-GCN combination demonstrated superior performance in predicting KM values. This indicates that combining different GNN architectures can enhance the model’s ability to capture complex molecular interactions.
Challenges and Future Directions
Despite the promising results, the researchers acknowledge certain limitations:
- Dataset Diversity: Ensuring comprehensive coverage of enzyme varieties in the training datasets is challenging. Limited data on less common enzymes may affect the model’s accuracy when predicting KM values for these enzymes.
- Model Interpretability: The complexity of the neural network architecture can make it difficult to understand precisely what the model has learned about enzyme-substrate interactions. This “black box” nature is a common critique in advanced machine learning applications.
- Potential Overfitting and Bias: Improvements are needed in refining model parameters to reduce the risk of overfitting, where the model performs well on training data but poorly on unseen data.
Significance of the Study
This research marks a significant step in applying advanced machine learning techniques to bioinformatics. By expediting the prediction of KM values, GraphKM can:
- Accelerate Drug Development: Quickly identifying enzyme kinetics can streamline the development of enzyme-targeting drugs.
- Enhance Biochemical Synthesis: Understanding enzyme behavior aids in designing more efficient industrial biocatalysts.
- Deepen Biological Understanding: Accurate KM predictions contribute to fundamental research in enzymology and metabolic pathways.
GraphKM represents an innovative fusion of deep learning and enzyme kinetics, offering a powerful tool to predict the Michaelis constant for a wide range of enzymes. While challenges remain in dataset diversity and model interpretability, the study opens doors to more efficient and cost-effective research in medicine, industrial biology, and beyond. As cross-disciplinary approaches continue to evolve, integrating advanced computing with molecular biology holds great promise for future scientific breakthroughs.
References
- Original Research Paper. GraphKM: Machine and Deep Learning for KM Prediction of Wildtype and Mutant Enzymes. BMC Bioinformatics.
Keywords
- GraphKM, Enzyme Kinetics, Michaelis Constant (KM), Deep Learning, Graph Neural Networks (GNN), PaddlePaddle Framework, Transformer-Based Language Model, Bioinformatics, Machine Learning in Biology, Enzyme-Substrate Interaction
Disclaimer: The information presented in this article is for informational purposes only and reflects the findings of the referenced study as of the publication date. For detailed information, please refer to the original research paper or consult professionals in the field.