Deep Learning Powers New Advances in Enzyme Engineering
Researchers from the University of Washington, led by Anna Lauko and Samuel J. Pellock, have developed a novel computational method for designing serine hydrolases from scratch. Detailed in their preprint titled “Computational Design of Serine Hydrolases“ on bioRxiv, this work involves a large team including senior author David Baker. The study represents a significant advancement in synthetic biology and enzyme engineering, utilizing deep learning tools to create enzymes with specific functions.
Challenges in Designing De Novo Enzymes
Designing enzymes from the ground up is a complex task due to the precise arrangement required in their active sites—the regions where chemical reactions occur. Traditional methods have struggled to replicate the efficiency and specificity of natural enzymes. The team’s approach addresses these challenges by employing advanced computational tools to model and predict enzyme structures with the necessary active site geometry for catalytic activity.
Innovative Computational Tools
The researchers utilized a deep learning method called RFdiffusion, which assists in creating proteins with specific active site configurations. This tool allows for the generation of protein structures that can accommodate the precise positioning of amino acids necessary for enzyme function. Additionally, they developed ChemNet, a computational model that predicts whether an enzyme’s active site is appropriately structured to function throughout the reaction cycle.
Experimental Results
Focusing on enzymes that catalyze ester hydrolysis, the team designed de novo enzymes incorporating a catalytic triad—a combination of serine, histidine, and aspartate residues essential for this reaction. They created multiple enzyme variants and tested them in the laboratory. Some of these designed enzymes successfully catalyzed the breakdown of esters, although their efficiency was lower compared to natural enzymes.
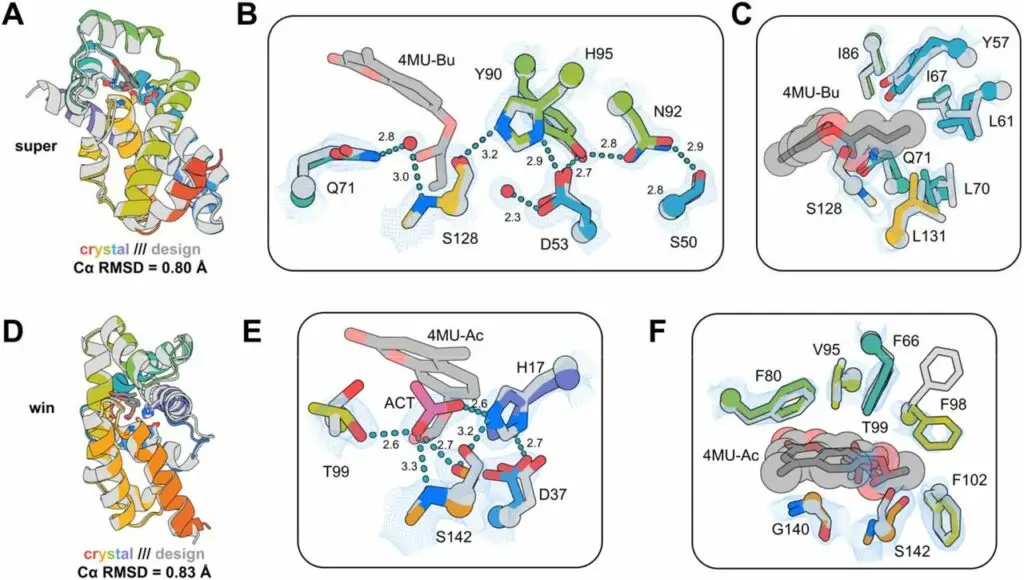
To verify the accuracy of their designs, the researchers employed X-ray crystallography to determine the three-dimensional structures of the enzymes. The experimental structures closely matched the predicted models, demonstrating the effectiveness of their computational methods in generating accurate enzyme structures.
Limitations and Future Directions
Despite the successful design and synthesis of functional enzymes, the engineered enzymes exhibited lower catalytic efficiency and stability compared to their natural counterparts. They often ceased functioning after catalyzing a few reactions. The team suggests that this may be due to suboptimal active site arrangements and incomplete optimization of the catalytic environment, such as the positioning of water molecules necessary for the reaction.
Further research is needed to enhance the catalytic efficiency and durability of these designed enzymes. Improving active site optimization and exploring additional computational techniques could lead to enzymes that match or surpass natural enzymes in performance.
Implications for Biotechnology
This study provides a framework for the de novo design of enzymes, which has significant implications for various fields:
- Industrial Processes: Custom-designed enzymes could improve the efficiency of chemical manufacturing and reduce reliance on harsh chemicals.
- Medicine: Enzymes tailored to specific reactions may lead to new therapies or diagnostic tools.
- Environmental Applications: Engineered enzymes could be developed to degrade pollutants or recycle materials more effectively.
The work by Lauko, Pellock, and colleagues represents a meaningful step forward in computational enzyme design. By integrating deep learning tools with experimental validation, they have demonstrated the potential to create functional enzymes from scratch. While challenges remain in enhancing the efficiency and stability of these enzymes, the study lays the groundwork for future advancements in enzyme engineering.
Reference
- Original Research Paper. Computational Design of Serine Hydrolases. bioRxiv. (Note: As this is a preprint, it has not yet undergone peer review.)
Keywords
De Novo Enzyme Design, Serine Hydrolases, Computational Biology, Deep Learning, RFdiffusion, ChemNet, Catalytic Triad, Ester Hydrolysis, Synthetic Biology, Enzyme Engineering
Disclaimer: The information presented in this article is based on a preprint study and has not undergone peer review. It is intended for informational purposes only. For more detailed information, please refer to the original preprint or consult professionals in the field.